3D visualisation and quantification of biofilm in porous media by using X-ray microtomography
Short term visitor's project
The biofilms, defined as structured communities of bacteria attached to a solid surface and surrounded by a protecting matrix of self-produced exopolymeric substances, are widely used in biotechnology for wastewater purification and biodegradation of xenobiotics. One of the ways to use biofilms is in the form of biofilters, where biofilms are grown and maintained on a granular porous media kept in a reactor under a constant flow. In this way the bacteria are protected from washout and remain in a more hospitable environment so the growth and treatment capacity are facilitated.
However, the biofilm processes are difficult to control because of the simultaneous interaction between the biofilm structure and the hydrodynamics of the flow; the liquid flow in the porous matrix determines the availability of nutrients and therefore the biomass growth and biofilm structure, while simultaneously, the growth of the biofilm modifies the liquid flow in the porous matrix. The development of a predictive model is thus a complex task but appears necessary to provide decision-aid tools for engineers and to improve the processes in which biofilms are involved.
So far, the models for biofilm growth in porous media are based on numerical modelling which draw experimental data from macro-scale (pilot biofilters, measuring of hydrodynamic parameters) and micro-scale (micro-fluidic chambers, confocal microscopy). But there is a lack of experimental data at the meso-scale; i.e. the scale of a few pores, which is the one relevant for industrial biofilters.
A new methodology based on X-ray microtomography giving information on the biofilm distribution and morphology at the pore size was developed recently in our labs and is at the basis of the present project. Thanks to the capacity of this imaging method to distinguish the biofilm structure from the surrounding liquid phase, this project aims at completing the characterisation and the quantification of the evolution of the 3D microstructure of a bacterial biofilm specifically grown in porous media and its coupling with a wide range of hydrodynamic conditions.
This characterization at the pore scale is necessary to develop macroscopic models that are as close as possible to reality.
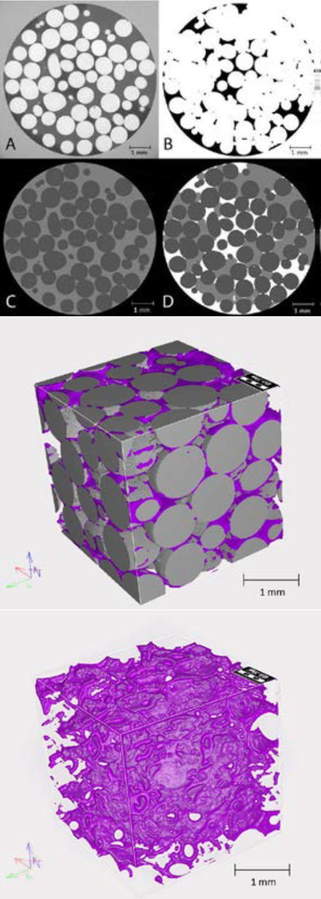
Figure caption: Segmentation of microtomographs by Fiji software; In the X-ray tomographs (A) different phases are in various shades of gray color which can be separated as biofilm (B) or beads (C). When images are merged (D) each phase is represented by a single color (biofilm=white, beads=dark gray, bulk liquid =light gray). Figure 2 (below): 3D model of biofilm constructed in Geodict program.
Presented are the images taken from columns filled with 1 mm glass beads and biofilm grown for 7 days at 21 ml h-1 flow rate.

CONTACTS
- PI: Tomislav Ivankovic (visitor)
- Co-PI: Sabine Rolland du Roscoat, Philippe Séchet, Zhujun Huang

PARTNERS
- Depatment of Microbiology (University fo Zagreb, Croatia)
- 3SR
- LEGI

FUNDING
Tec21
