3D mechanics of cells in complex fibrous media
PhD project
Cells are usually studied on 2D substrates and processes used by cells to migrate are rather well known. The stresses developed by cells on 2D functionalised substrates have been measured using the displacements of fluorescent beads embedded in the substrate. We have carried out this kind of analysis using an original method based on Finite Elements and proper regularisation tools, to solve the inverse problem with known displacements giving access to stresses at the cell-substrate interface (Fig. 1). This is also called Traction Force Microscopy (TFM).

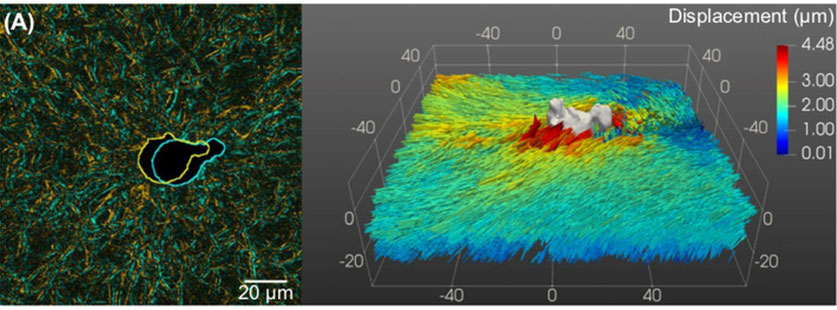
In 3D, it is more difficult to solve such problems but there have been some attempts using continuous modelling with elastic isotropic substrates. Our purpose is here to solve the case of a cell with complex shape in 3D anisotropic and viscohyperelastic media. This is a difficult inverse problem. Finally, numerical resolution can be costly. To determine the matrix displacements, we propose to use the fluorescence naturally reflected from fibres such as collagen, using a confocal microscope. This allows to measure displacement fields (Fig. 2) and thus proper finite strain fields within the extra-cellular matrix, made of amorphous ground substances and collagen fibrous networks. From this type of 3D images, it is also possible to extract relevant microstructure descriptors such as the extra-cellular matrix fibres waviness and orientation distribution, and to follow their evolution upon loading. The next step will be to estimate the stress states exerted by cells on the fibrous extra-cellular matrix. We choose here to use a micromechanical analysis to better understand what is occurring at the microscopic level (i.e. the level of fibres) and how such elementary forces combine to exert a given stress, or how do fibres resist stresses exerted by cells. Thanks to kinematical and structural data extracted from 4D experiments, and insights from molecular dynamics simulations, a proper micromechanical analysis will be carried out with relevance.
CONTACTS
Valérie Laurent (Project PI)
Lucie Bailly (co-PI)
Anaïs Reboullet (PhD student)
PARTNERS
LIPhy
3SR
